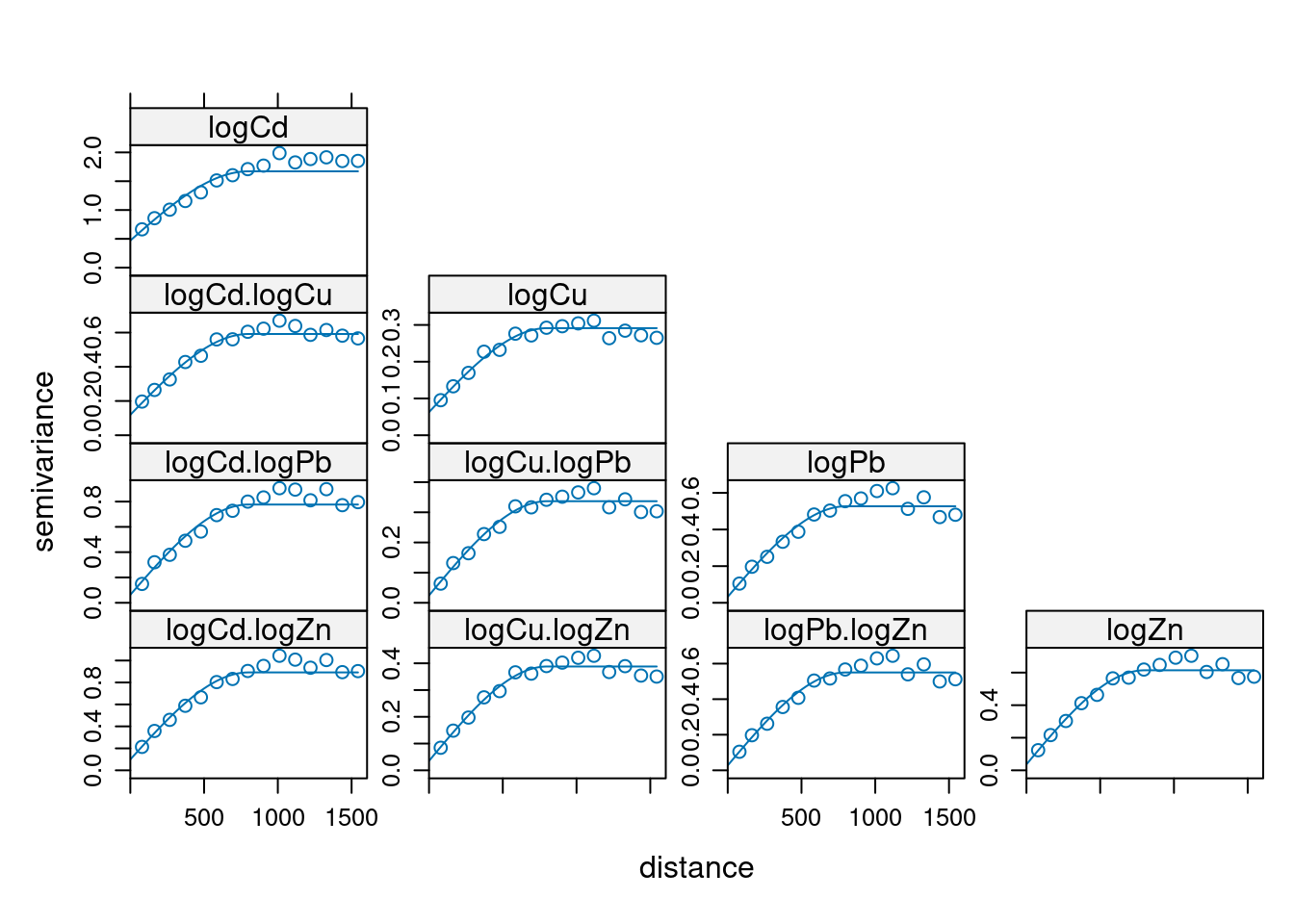
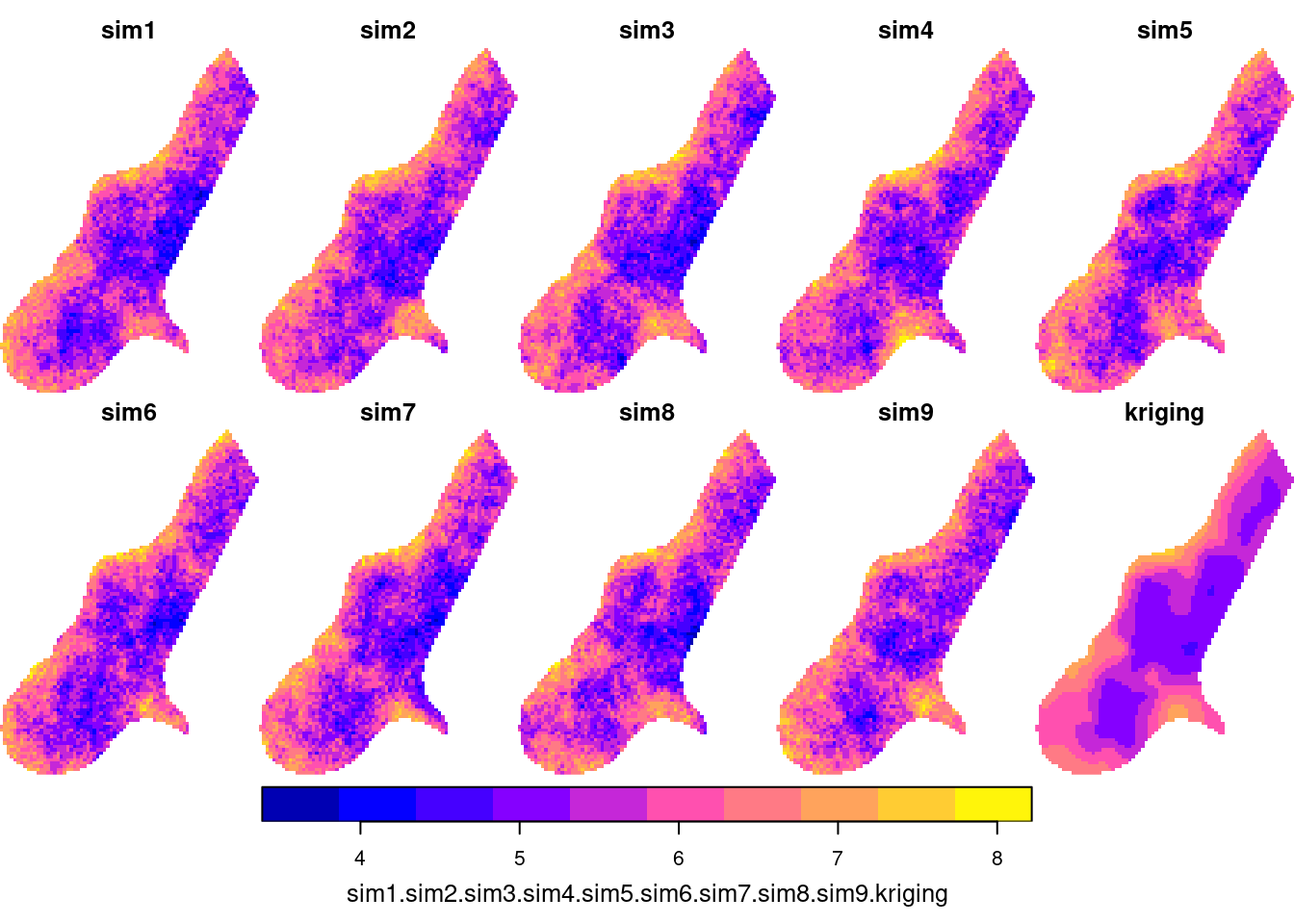
Cokriging sets the multivariate equivalent of kriging, which is, in terms of number of dependent variables, univariate. Kriging: \[Z(s) = X(s)\beta + e(s)\] Cokriging: \[Z_1(s) = X_1(s)\beta_1 + e_1(s)\] \[Z_2(s) = X_2(s)\beta_2 + e_2(s)\] \[Z_k(s) = X_k(s)\beta_k + e_k(s)\] with \(V = \Cov(e_1,e_2,...,e_k)\)
Cokriging prediction
Cokriging prediction is not substantially different from kriging prediction, it is just a lot of book-keeping.
How to set up \(Z(s)\), \(X\), \(\beta\), \(e(s)\), \(x(s_0)\), \(v\), \(V\)?
Multivariable prediction involves the joint prediction of multiple, both spatially and cross-variable correlated variables. Consider \(m\) distinct variables, and let \(\{Z_i(s), X_i, \beta^i, e_i(s), x_i(s_0), v_i, V_i\}\) correspond to \(\{Z(s), X, \beta, e(s), x(s_0), v, V\}\) of the \(i\)-th variable. Next, let \({\bf Z}(s) = (Z_1(s)',...,Z_m(s)')'\), \({\bf B}=({\beta^1} ',...,{\beta^m} ')'\), \({\bf e}(s)=(e_1(s)',...,e_m(s)')'\),
\[
{\bf X} =
\left[
\begin{array}{cccc}
X_1 & 0 & ... & 0 \\\\
0 & X_2 & ... & 0 \\\\
\vdots & \vdots & \ddots & \vdots \\\\
0 & 0 & ... & X_m \\\\
\end{array}
\right], \
{\bf x}(s_0) =
\left[
\begin{array}{cccc}
x_1(s_0) & 0 & ... & 0 \\\\
0 & x_2(s_0) & ... & 0 \\\\
\vdots & \vdots & \ddots & \vdots \\\\
0 & 0 & ... & x_m(s_0) \\\\
\end{array}
\right]
\]
with \(0\) conforming zero matrices, and
\[{\bf v} =
\left[
\begin{array}{cccc}
v_{1,1} & v_{1,2} & ... & v_{1,m} \\\\
v_{2,1} & v_{2,2} & ... & v_{2,m} \\\\
\vdots & \vdots & \ddots & \vdots \\\\
v_{m,1} & v_{m,2} & ... & v_{m,m} \\\\
\end{array}
\right], \ \
{\bf V} =
\left[
\begin{array}{cccc}
V_{1,1} & V_{1,2} & ... & V_{1,m} \\\\
V_{2,1} & V_{2,2} & ... & V_{2,m} \\\\
\vdots & \vdots & \ddots & \vdots \\\\
V_{m,1} & V_{m,2} & ... & V_{m,m} \\\\
\end{array}
\right]
\]
where element \(i\) of \(v_{k,l}\) is \(\Cov(Z_k(s_i), Z_l(s_0))\), and where element \((i,j)\) of \(V_{k,l}\) is \(\Cov(Z_k(s_i),Z_l(s_j))\).
The multivariable prediction equations equal the previous UK equations and when all matrices are substituted by their multivariable forms (see also Ver Hoef and Cressie, Math.Geol., 1993), and when for \(\sigma^2_0\), \(\Sigma\) is substituted with \(\Cov(Z_i(s_0),Z_j(s_0))\) in its \((i,j)\)-th element. Note that the prediction variance is now a prediction error covariance matrix.